SKLRD & Shenzhen Huada Gene Research Institute in collaboration to analyze the cfRNA characteristics of critical COVID-19 patients
2022-01-271291On January 21, 2022, the team led by Zhao Jincun with SKLRD and the joint research team of Shenzhen Huada Life Science Institute published a research paper titled “Plasma cell-free RNA characteristics in COVID-19 patients” in the international famous academic journal Genome Research1. By comparing the differences of plasma cell-free RNA (cfRNA) between patients with different symptoms of COVID-19 and healthy controls, a series of genes differentially expressed in patients with severe COVID-19 have been found, and a gene regulatory network that may lead to cytokine storm has been found accordingly, providing new ideas for analyzing the pathogenesis of severe cases of COVID-19.

Figure 1. First page of the paper published in the official website of Genome Research
Plasma cfRNA carries unique information from the different tissues and organs of the human body, and can provide an important reference for studying the disease mechanisms and the host-pathogen interactions. The study used the PALM-Seq technology independently developed by the Huada Life Science Institute, and also tested the full spectrum of cell-free mRNA, lncRNA, miRNA and tRNA in the plasma of patients with different symptoms and healthy controls. A total of seven biomarkers associated with severe COVID-19 were identified, and the neutrophil activation pathway in severe patients was activated. It was found through lncRNA analysis that the translation level of IL-6R was enhanced in COVID-19 patients, and based on the data, it proved that the enhancement was achieved by down-regulating miRNA such as miR-451a and let-7, further validating the previous work results of the team, and revealing the potential regulatory mechanism of miR-451a and the corresponding lncRNA on the inflammatory factor-mediated storm of IL-6 / IL-6R among COVID-19 patients2.
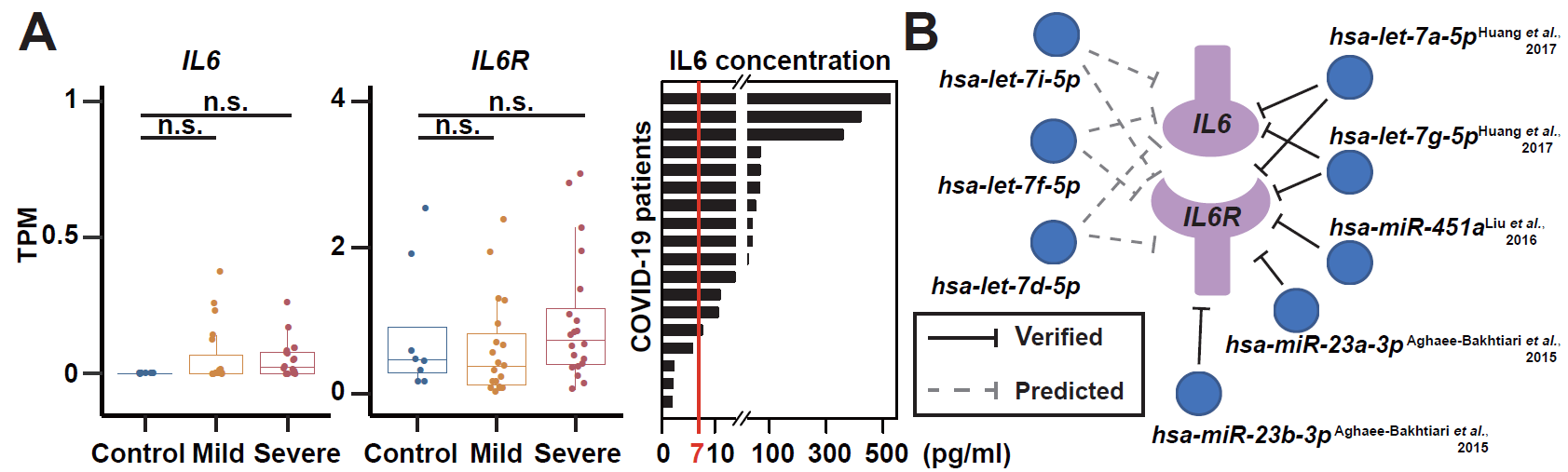
Figure 2. The targeting relationship between miRNA and IL-6 / IL-6R
Meanwhile, the study also found that the expression levels of tRNA molecules were up-regulated in COVID-19 patients. The overall abundance of tRNA molecules and codon abundance after conversion to the corresponding codons showed a high consistency between mild and severe cases. The results of Principal component analysis (PCA) showed that codon abundance can be used to distinguish COVID-19 patients from healthy controls. Subsequent codon preference analysis indicates that these up-regulated codons are more likely to promote the translation of novel coronavirus mRNA, and these codons end with A and T, which is consistent with novel coronavirus codon preference for the ending of A / T. The feature is expected to become a specific target of novel coronavirus drugs and vaccines in the future.
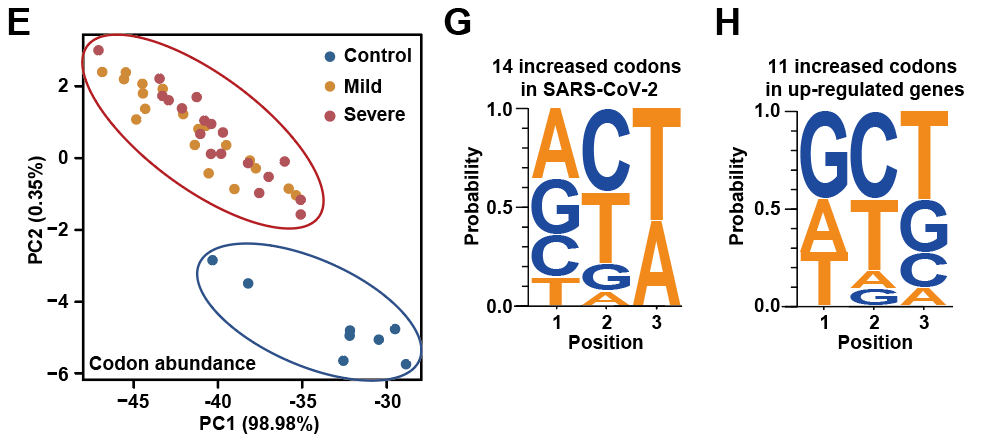
Figure 3. Novel Coronavirus codon preference for the ending of A / T
The study also found that other pneumonia-causing microorganisms can be detected in the plasma cell-free RNA in COVID-19 patients, which also suggests that we should pay attention to the co-infection phenomenon in the lungs of COVID-19 patients.
References:
1. Wang, Yanqun, Jie Li, Lu Zhang, Hai-Xi Sun, Zhaoyong Zhang, Jinjin Xu, Yonghao Xu et al. "Plasma cell-free RNA characteristics in COVID-19 patients." Genome Research (2022).
2. Yang, Penghui, Yingze Zhao, Jie Li, Chuanyu Liu, Linnan Zhu, Jie Zhang, Yeya Yu et al. "Downregulated miR-451a as a feature of the plasma cfRNA landscape reveals regulatory networks of IL-6/IL-6R-associated cytokine storms in COVID-19 patients." Cellular & Molecular Immunology 18, no. 4 (2021): 1064-1066.
Original paper:https://genome.cshlp.org/content/early/2022/01/21/gr.276175.121.long
















