To Establish a Co-Infection Model of Influenza viruses and Bacteria, Results Shown in Multi-Disciplinary Interaction
2021-07-15881To Establish a Co-Infection Model of Influenza viruses and Bacteria, Results Shown in Multi-Disciplinary InteractionInfluenza, caused by influenza virus, is one of the commonest infectious diseases that affect human health. According to conservative estimates, in the 1918 influenza pandemic that caused more than 50 million deaths worldwide, 90% of the deaths were caused by influenza viruses that induced bacterial co-infection, rather than the direct effects of viruses. Similarly, during the 2009 H1N1 influenza pandemic, bacterial co-infection was positively correlated with the mortality caused by H1N1, and 18%-34% of ICU patients had bacterial co-infection. Streptococcus pneumoniae, Staphylococcus aureus, haemophilus influenzae, moraxella catarrhalis, etc. are common co-infective bacteria of influenza. Although antibiotics and influenza vaccines were successively introduced after 1918, death caused by viral and bacterial infections is still an important issue.

Professor Yang Zifeng’s team from the lab is devoted to the study on the diagnosis and treatment of respiratory viruses, giving full play to the combination of basic and clinical study. Through the multi-disciplinary researches of viruses, pharmacology, and clinics, the team continued to explore the research direction of influenza virus and bacterial co-infection, achieving important results especially in the establishment of models of influenza virus and bacterial co-infection, laying a foundation for the study of follow-up mechanisms and preventive strategies.
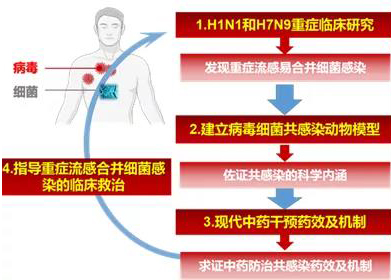
Figure 1 The clinical-model-drug co-infection research cycle
In the severe clinical study of H1N1 flu, Professor Yang Zifeng’s research team found that H1N1 virus infection can activate genes related to chemotaxis and cellular motility, making it easier for opportunistic pathogens to colonize and co-infect the lower respiratory tract [1]. In the follow-up study of H7N9, a new transcriptomic analysis method (MDTH, Molecular distance to health) was developed to determine the key host response and transcription characteristics closely related to the clinic, proving that the MDTH in patients with critical H7N9 is related to the bacterial infection and viral load of the lower respiratory tract for the first time. It was further found that 8 innate immune regulatory genes like CD177, MMP9, and MGAM2 are closely related to severe bacterial co-infection, confirming the view that severe avian influenza is more likely to be associated with bacterial infection [2].
Due to the limited and difficult clinical samples, in order to deeply study the mechanism of occurrence and development, drug screening and treatment evaluation of co-bacterial infection of influenza virus. According to common co-infecting bacteria of influenza, Yang Zifeng’s team successively established the mouse model for secondary staphylococcus aureus (SA) of influenza virus [3], secondary methicillin-resistant staphylococcus aureus (MRSA) [4] and secondary haemophilus influenzae (NTHi) [5]. Compared with single-factor viral or bacterial co-infections, these co-infection models perform more obviously in lung inflammation and pathological damage, and the mortality rate also increased significantly. showing homology of pathogenesis and consistent behavioral appearance, as well as ease of operation and economic feasibility.
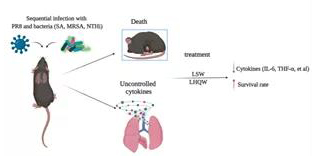
Figure 2 Schematic diagram of co-infection model establishment and drug intervention
With the co-infection mouse model, Professor Yang Zifeng’s team conducted a preliminary exploration of drug intervention and related mechanisms, and found that influenza virus infection can promote the expression of adhesion molecules, which has a positive correlation with bacterial titer in lungs, suggesting that adhesion molecules may play a certain role in the influenza virus-induced bacterial infection. The traditional Chinese medicine Lianhua Qingwen and Liushen Wan were used to intervene the co-infection models of influenza virus and secondary staphylococcus aureus respectively, and found that they have a protective effect on severe pneumonia caused by co-infection, which can significantly heal lung pathological damage and reduce lung inflammation. In particular, Liushen Wan can also extend the survival of co-infected mice; and its protective mechanism is likely to be achieved by inhibiting the up-regulation of adhesion molecules, thereby inhibiting the adhesion of bacteria after influenza virus infection. On the basis of three different co-infection animal models, the team will further explore the mechanism of co-infection occurrence and development, as well as the evaluation of drug intervention effects, in order to provide a basis for clinical prevention, treatment and drug selection of severe influenza.
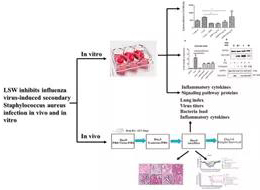
Figure 3 Liushen Wan inhibits secondary staphylococcus aureus of influenza virus
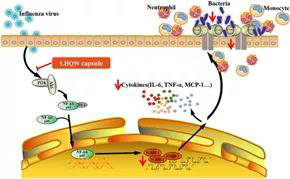
Figure 4 Lianhua Qingwen inhibits MRSA infection
















